In bioinformatics, the comparison and alignment of biological sequences is very important. From understanding the genetic codes that define life to identifying functional domains and conserved regions. The process of alignment involves pairing two or more sequences to discover their similarities, differences, and evolutionary relationships.
Global Sequence Alignment seeks to find the best alignment of entire sequences from the beginning to the end, emphasizing overall similarity even at the cost of introducing gaps at either terminus. Local Sequence Alignment concentrates on discovering regions of high similarity, accommodating gaps within the sequences and permitting the identification of conserved motifs and functional domains.
What Is Sequence Alignment?
A sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the distance cost between strings in a natural language or in financial data.
Global Sequence Alignment
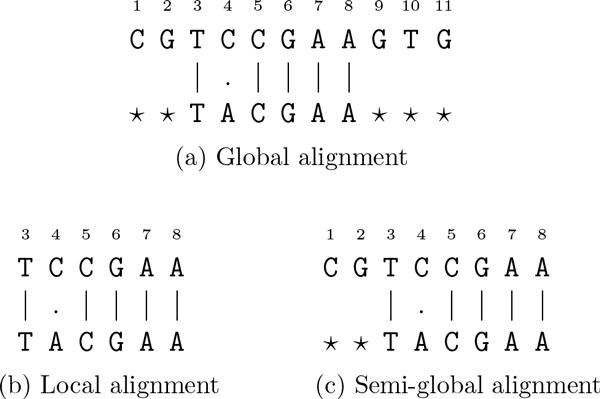
Global alignment is used to compare sequences in cases where we have reason to believe that the sequences are related along their entire length. If for example, sequences s and t are two independent sequencing runs of the same PCR product, then they should differ only in positions where there are sequencing errors. In order to find those sequencing errors, we align all of sequence s with all of sequence t. Other applications of global alignment include finding mutations in closely related gene or protein sequences and identification of single nucleotide polymorphisms (SNPs).
Examples of Global alignment tools include:
- EMBOSS Needle
- Needleman-Wunsch Global Align Nucleotide Sequences (Specialized BLAST)
Local Sequence Alignment
Local alignment addresses cases where we only expect to find isolated regions of similarity. One example is alignment of genomic DNA upstream from two co-expressed genes to find conserved regions that may correspond to transcription factor binding sites. Another application is identification of conserved domains1 in two amino acid sequences that encode proteins that share one or more domains, but are otherwise unrelated.
Examples of Local alignment tools include:
- BLAST
- EMBOSS Water
- LALIGN
Also Read: Difference Between Replication And Transcription
Difference Between Global And Local Sequence Alignment In Tabular Form
| BASIS OF COMPARISON | GLOBAL SEQUENCE ALIGNMENT | LOCAL SEQUENCE ALIGNMENT |
| Description | In global alignment, an attempt is made to align the entire sequence (end to end alignment). | Finds local regions with the highest level of similarity between the two sequences. |
| Examples Of Tools | -EMBOSS Needle -Needleman-Wunsch Global Align Nucleotide Sequences (Specialized BLAST) | -BLAST -EMBOSS Water -LALIGN |
| Function | A global alignment contains all letters from both the query and target sequences. | A local alignment aligns a substring of the query sequence to a substring of the target sequence. |
| Two Sequences | If two sequences have approximately the same length and are quite similar, they are suitable for global alignment. | Any two sequences can be locally aligned as local alignment finds stretches of sequences with high level of matches without considering the alignment of rest of the sequence regions. |
| Suitability | Suitable for aligning two closely related sequences. | Suitable for aligning more divergent sequences or distantly related sequences. |
| Use | Global alignments are usually done for comparing homologous genes like comparing two genes with same function (in human vs. mouse) or comparing two proteins with similar function. | Used for finding out conserved patterns in DNA sequences or conserved domains or motifs in two proteins. |
| General Technique | A general global alignment technique is the Needleman–Wunsch algorithm. | A general local alignment method is Smith–Waterman algorithm. |
Also Read: Difference Between Pairwise And Multiple Sequence Alignment
Key Takeaways
Purpose
- Global alignment is suitable for comparing sequences with significant overall similarity and evolutionary relationships. It is commonly used when comparing homologous sequences that share a common ancestor.
- Local alignment is useful for identifying short conserved regions or motifs within sequences, such as functional domains or similar regions that may have undergone convergent evolution.
Algorithm used
- The Needleman-Wunsch algorithm is commonly used for global sequence alignment. It employs dynamic programming to find the optimal alignment between two sequences.
- The Smith-Waterman algorithm is the standard approach for local sequence alignment. Like Needleman-Wunsch, it also utilizes dynamic programming but with the added feature of allowing negative scores to handle local alignments.
Scoring system
- In global alignment, the scoring system generally includes positive scores for matching residues and negative scores for mismatches and gaps. The aim is to maximize the total score of the alignment.
- Local alignment utilizes a similar scoring scheme as global alignment, but it allows the score to drop below zero for regions of low similarity, which is crucial for detecting local similarities.
Usage of gaps
- Global alignment introduces gaps at the beginning and end of sequences to ensure alignment across their entire lengths, even if it results in lower local similarity scores.
- Local alignment permits gaps only within the sequence regions being aligned, which allows for the identification of local regions of similarity without requiring alignment across the entire sequences.
Output
- The output of global alignment is the entire alignment of both sequences, providing a complete picture of their similarity across their full lengths.
- The output of local alignment is the alignment of the best-matching subsequences, highlighting regions of high similarity within the sequences.
Use cases
- Global alignment is commonly used for evolutionary studies, phylogenetic analysis, and comparing highly similar sequences across species.
- Local alignment is useful for identifying functional similarities or conserved regions within related sequences, finding short sequence motifs, and characterizing potential functional domains.